susie_plot produces a per-variable summary of
the SuSiE credible sets. susie_plot_iteration produces a
diagnostic plot for the susie model fitting. For
susie_plot_iteration, several plots will be created if
track_fit = TRUE when calling susie.
susie_plot(
model,
y,
add_bar = FALSE,
pos = NULL,
b = NULL,
max_cs = 400,
add_legend = NULL,
...
)
susie_plot_iteration(model, L, file_prefix, pos = NULL)Arguments
- model
A SuSiE fit, typically an output from
susieor one of its variants. Forsuse_plot, the susie fit must havemodel$z,model$PIP, and may includemodel$sets.modelmay also be a vector of z-scores or PIPs.- y
A string indicating what to plot: either
"z_original"for z-scores,"z"for z-score derived p-values on (base-10) log-scale,"PIP"for posterior inclusion probabilities,"log10PIP"for posterior inclusion probabiliities on the (base-10) log-scale. For any other setting, the data are plotted as is.- add_bar
If
add_bar = TRUE, add horizontal bar to signals in credible interval.- pos
Indices of variables to plot. If
pos = NULLall variables are plotted.- b
For simulated data, set
b = TRUEto highlight "true" effects (highlights in red).- max_cs
The largest credible set to display, either based on purity (set
max_csbetween 0 and 1), or based on size (setmax_cs > 1).- add_legend
If
add_legend = TRUE, add a legend to annotate the size and purity of each CS discovered. It can also be specified as location where legends should be added, e.g.,add_legend = "bottomright"(default location is"topright").- ...
Additional arguments passed to
plot.- L
An integer specifying the number of credible sets to plot.
- file_prefix
Prefix to path of output plot file. If not specified, the plot, or plots, will be saved to a temporary directory generated using
tempdir.
Value
Invisibly returns NULL.
See also
Examples
set.seed(1)
n = 1000
p = 1000
beta = rep(0,p)
beta[sample(1:1000,4)] = 1
X = matrix(rnorm(n*p),nrow = n,ncol = p)
X = scale(X,center = TRUE,scale = TRUE)
y = drop(X %*% beta + rnorm(n))
res = susie(X,y,L = 10)
susie_plot(res,"PIP")
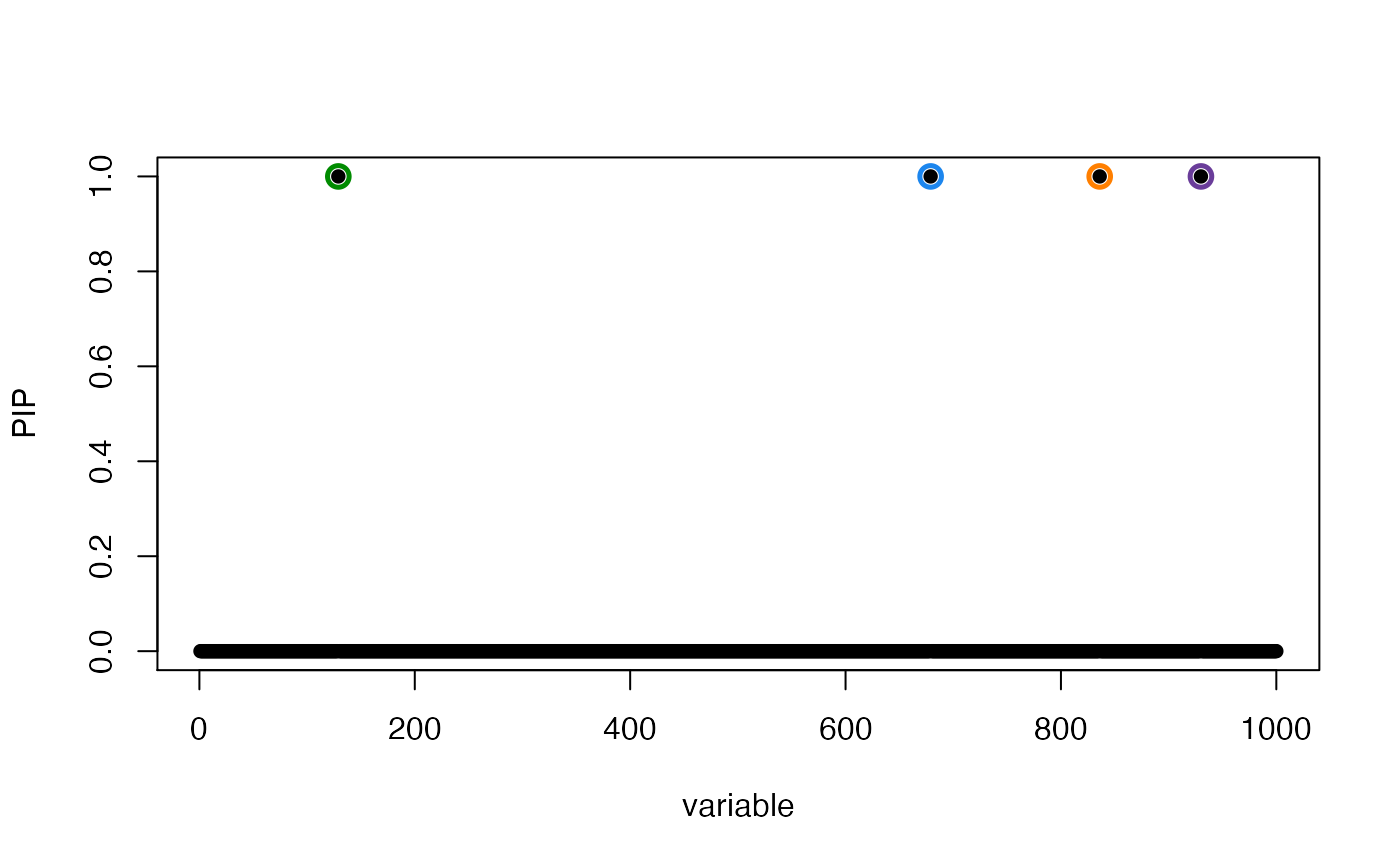 susie_plot(res,"PIP",add_bar = TRUE)
susie_plot(res,"PIP",add_bar = TRUE)
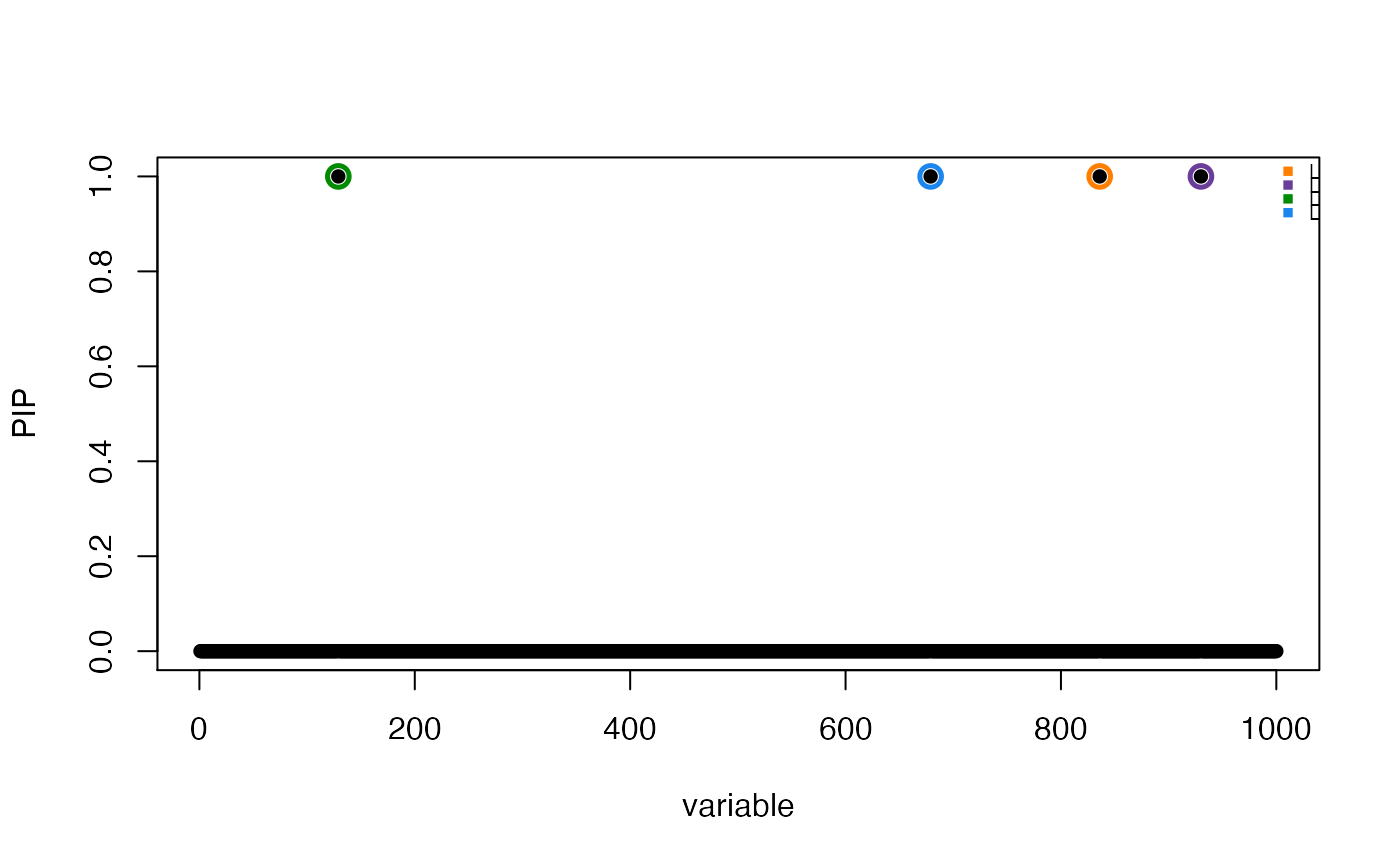
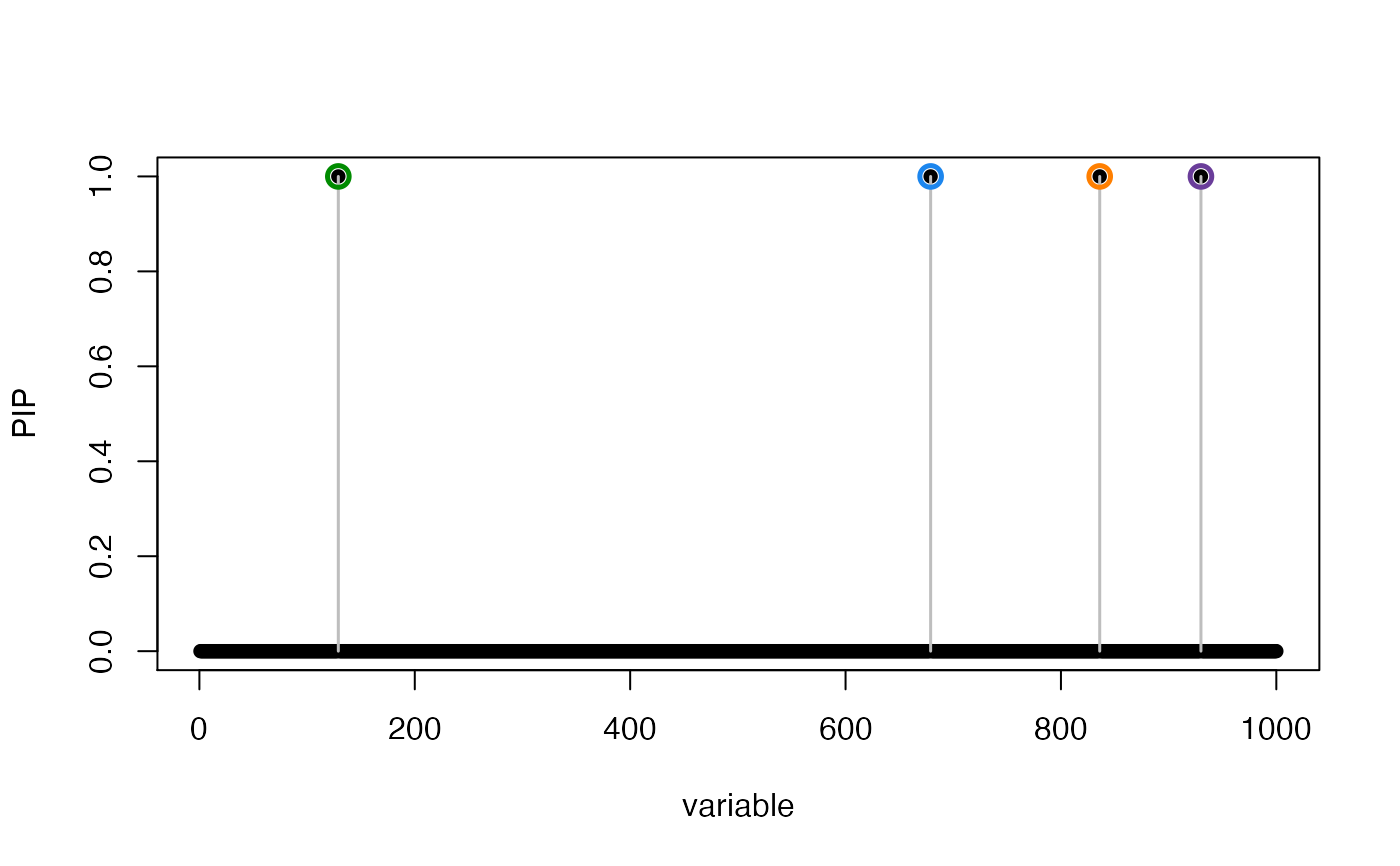 susie_plot(res,"PIP",add_legend = TRUE)
susie_plot(res,"PIP",add_legend = TRUE)
 susie_plot(res,"PIP", pos=1:500, add_legend = TRUE)
susie_plot(res,"PIP", pos=1:500, add_legend = TRUE)
 # Plot selected regions with adjusted x-axis position label
res$genomic_position = 1000 + (1:length(res$pip))
susie_plot(res,"PIP",add_legend = TRUE,
pos = list(attr = "genomic_position",start = 1000,end = 1500))
# Plot selected regions with adjusted x-axis position label
res$genomic_position = 1000 + (1:length(res$pip))
susie_plot(res,"PIP",add_legend = TRUE,
pos = list(attr = "genomic_position",start = 1000,end = 1500))
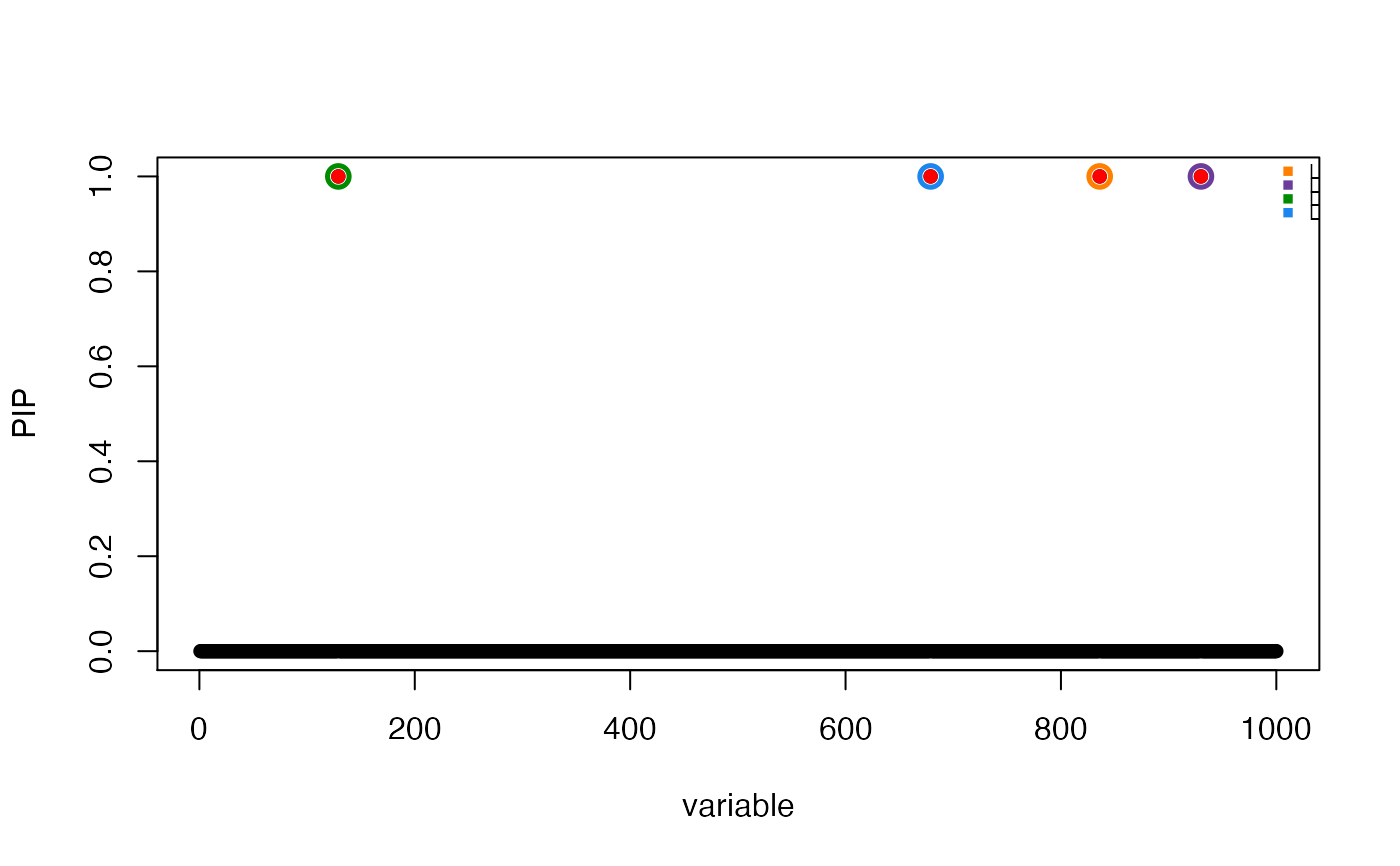
 # True effects are shown in red.
susie_plot(res,"PIP",b = beta,add_legend = TRUE)
# True effects are shown in red.
susie_plot(res,"PIP",b = beta,add_legend = TRUE)
 set.seed(1)
n = 1000
p = 1000
beta = rep(0,p)
beta[sample(1:1000,4)] = 1
X = matrix(rnorm(n*p),nrow = n,ncol = p)
X = scale(X,center = TRUE,scale = TRUE)
y = drop(X %*% beta + rnorm(n))
res = susie(X,y,L = 10)
susie_plot_iteration(res, L=10)
#> Iterplot saved to /var/folders/9b/ck4lp8s140lcksryyh4dppdr0000gn/T//Rtmpj3YNjC/susie_plot.pdf
set.seed(1)
n = 1000
p = 1000
beta = rep(0,p)
beta[sample(1:1000,4)] = 1
X = matrix(rnorm(n*p),nrow = n,ncol = p)
X = scale(X,center = TRUE,scale = TRUE)
y = drop(X %*% beta + rnorm(n))
res = susie(X,y,L = 10)
susie_plot_iteration(res, L=10)
#> Iterplot saved to /var/folders/9b/ck4lp8s140lcksryyh4dppdr0000gn/T//Rtmpj3YNjC/susie_plot.pdf