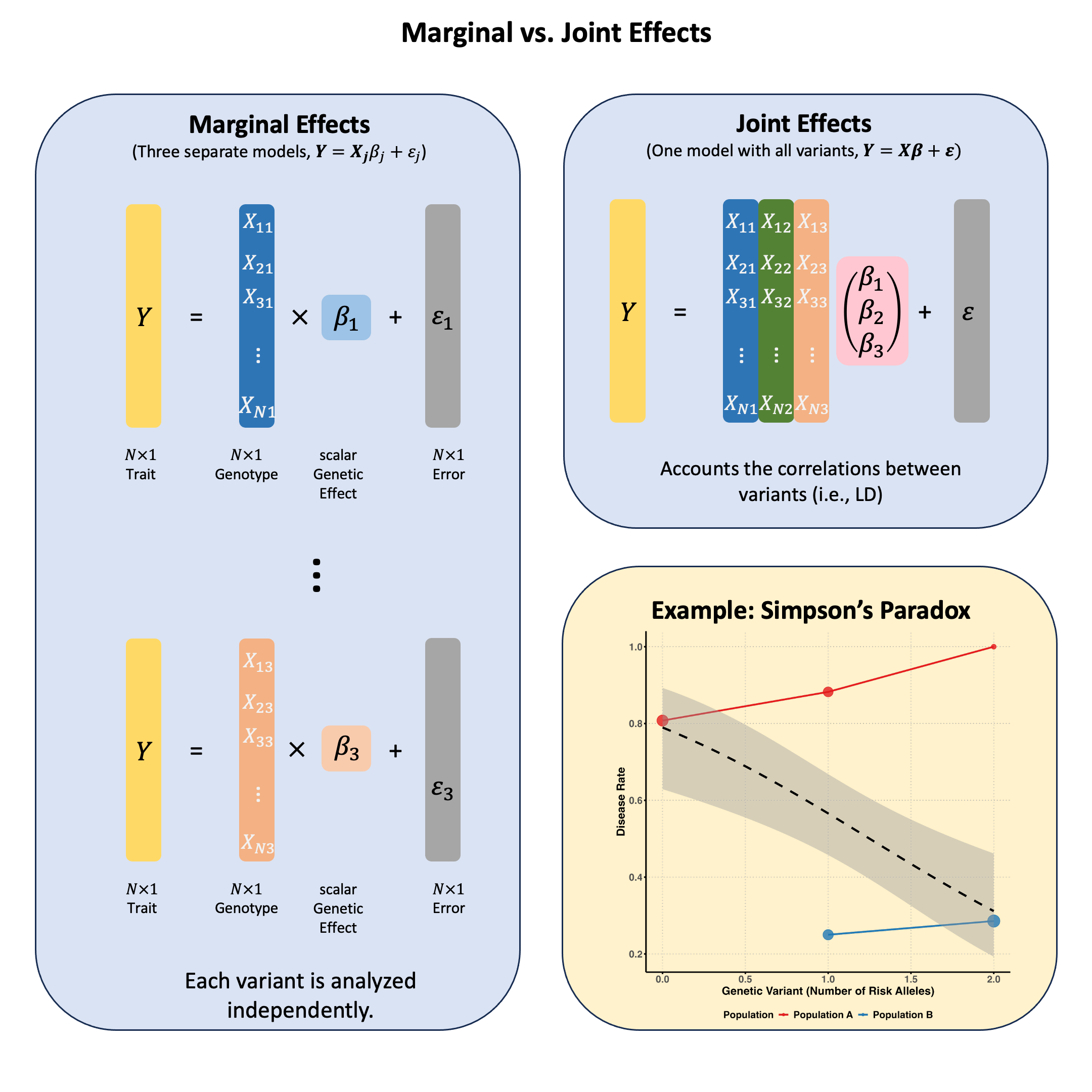
Marginal and Joint Effects#
Marginal effects measure a genetic variant’s influence on a trait when considered alone, ignoring other variants, while joint effects measure each variant’s independent contribution when all variants are simultaneously included in the model, revealing their true effects after accounting for correlations (LD) between them.
Graphical Summary#
Key Formula#
In multiple markers linear regression, we extend the single marker model to incorporate multiple genetic variants:
Where:
\(\mathbf{Y}\) is the \(N \times 1\) vector of trait values for \(N\) individuals
\(\mathbf{X}\) is the \(N \times M\) matrix of genotypes for \(M\) variants across \(N\) individuals
\(\boldsymbol{\beta}\) is the \(M \times 1\) vector of effect sizes for each variant (to be estimated)
\(\boldsymbol{\epsilon}\) is the \(N \times 1\) vector of error terms for \(N\) individuals and \(\boldsymbol{\epsilon} \sim N(0, \sigma^2\mathbf{I})\)
We can still use ordinary least squares (OLS) to derive the estimators for \(\boldsymbol{\beta}\) in matrix form:
Technical Details#
Marginal Effect#
In OLS, we discuss the single marker linear regression, which actually estimates the marginal effect of each genetic variant, considering each variant can be treated independently.
The marginal effect of a genetic variant is its association with the trait when analyzed in isolation, without accounting for other variants:
Where \(\mathbf{X}_j\) is the column vector for the \(j\)-th variant.
Joint Effect#
The joint effect of a genetic variant is its association with the trait when analyzed simultaneously with other variants, i.e., in the multiple markers model:
Where \(\hat{\beta}_{\text{joint},j}\) (the \(j\)-th element of \(\hat{\boldsymbol{\beta}}_{\text{joint}}\)) represents the effect of the \(j\)-th variant after accounting for all other variants in the model.
Key Differences Between Marginal and Joint Effects#
Correlation Structure:
Marginal effects ignore correlations (linkage disequilibrium) between variants
Joint effects account for correlations between variants
Interpretation:
Marginal effect: The expected change in the trait associated with a unit change in the variant, not accounting for other variants
Joint effect: The expected change in the trait associated with a unit change in the variant, holding all other variants constant
Consistency:
When variants are uncorrelated, marginal and joint effects are identical (\(\hat{\beta}_{\text{marginal}} = \hat{\beta}_{\text{joint}}\)), but joint model still provides advantages:
Better precision: More accurate standard errors from pooled residual variance estimation
Reduced multiple testing: One coherent test instead of many separate tests requiring correction
When variants are correlated, marginal and joint effects will differ
Joint effects can be smaller or larger than marginal effects, or even have opposite signs
Example#
Example 1 – Causal Variant Identification with LD#
When testing genetic variants individually, we might find that multiple variants appear to be associated with a trait. But what happens when we test them together? This example shows how linkage disequilibrium (correlation between nearby variants) can make non-causal variants appear significant in marginal analysis, while joint analysis reveals which variant is truly driving the association.
The key question: Which genetic variants are truly causal versus just correlated with the causal variant?
# Clear the environment
rm(list = ls())
set.seed(9) # For reproducibility
# Define genotypes for 10 individuals at 3 variants
# Create correlated genotypes to simulate linkage disequilibrium
N = 20
M = 3
# Generate correlated genotype data
# Variant 1 is the true causal variant
# Variants 2 and 3 are in LD with variant 1
variant1 <- sample(0:2, N, replace = TRUE, prob = c(0.4, 0.4, 0.2))
# Create LD: variants 2 and 3 are correlated with variant 1
variant2 <- ifelse(runif(N) < 0.9, variant1, sample(0:2, N, replace = TRUE))
variant3 <- ifelse(runif(N) < 0.8, variant1, sample(0:2, N, replace = TRUE))
# Combine into matrix
Xraw_additive <- cbind(variant1, variant2, variant3)
rownames(Xraw_additive) <- paste("Individual", 1:N)
colnames(Xraw_additive) <- paste("Variant", 1:M)
# Standardize genotypes
X <- scale(Xraw_additive, center = TRUE, scale = TRUE)
# Generate phenotype where only Variant 1 has a true causal effect
true_beta1 <- 1.5 # Strong effect for variant 1
epsilon <- rnorm(N, mean = 0, sd = 0.5)
Y_raw <- X[, 1] * true_beta1 + epsilon # Only variant 1 affects the trait
# Standardize phenotype
Y <- scale(Y_raw)
We first recall how we calculate the marginal effect of each variant in OLS:
# Calculate marginal effects (one SNP at a time)
p_values <- numeric(M) # Store p-values
betas <- numeric(M) # Store estimated effect sizes
for (j in 1:M) {
SNP <- X[, j] # Extract genotype for SNP j
model <- lm(Y ~ SNP) # OLS regression: Trait ~ SNP
summary_model <- summary(model)
# Store p-value and effect size (coefficient)
p_values[j] <- summary_model$coefficients[2, 4] # p-value for SNP effect
betas[j] <- summary_model$coefficients[2, 1] # Estimated beta coefficient
}
marginal_OLS_results <- data.frame(
Variant = colnames(X),
Beta = round(betas, 4),
P_Value = round(p_values, 4),
Significant = p_values < 0.05
)
Now let’s calculate the joint effects by including all variants in one model:
# Multiple regression model including all variants simultaneously
joint_model <- lm(Y ~ X)
joint_summary <- summary(joint_model)
# Extract the joint effect coefficients and p-values
# Skip the intercept (first row)
joint_betas <- joint_summary$coefficients[2:(M+1), 1]
joint_p_values <- joint_summary$coefficients[2:(M+1), 4]
# Create results table for joint effects
joint_OLS_results <- data.frame(
Variant = colnames(X),
Beta = round(joint_betas, 4),
P_Value = round(joint_p_values, 4),
Significant = joint_p_values < 0.05
)
rownames(joint_OLS_results) <- NULL
print("Marginal Effect:")
marginal_OLS_results
print("Joint Effect:")
joint_OLS_results
[1] "Marginal Effect:"
| Variant | Beta | P_Value | Significant |
|---|---|---|---|
| <chr> | <dbl> | <dbl> | <lgl> |
| Variant 1 | 0.9416 | 0.0000 | TRUE |
| Variant 2 | 0.4997 | 0.0249 | TRUE |
| Variant 3 | 0.8226 | 0.0000 | TRUE |
[1] "Joint Effect:"
| Variant | Beta | P_Value | Significant |
|---|---|---|---|
| <chr> | <dbl> | <dbl> | <lgl> |
| Variant 1 | 1.0253 | 0.0001 | TRUE |
| Variant 2 | 0.0757 | 0.4226 | FALSE |
| Variant 3 | -0.1323 | 0.4886 | FALSE |
Example 2 – Simpson’s Paradox#
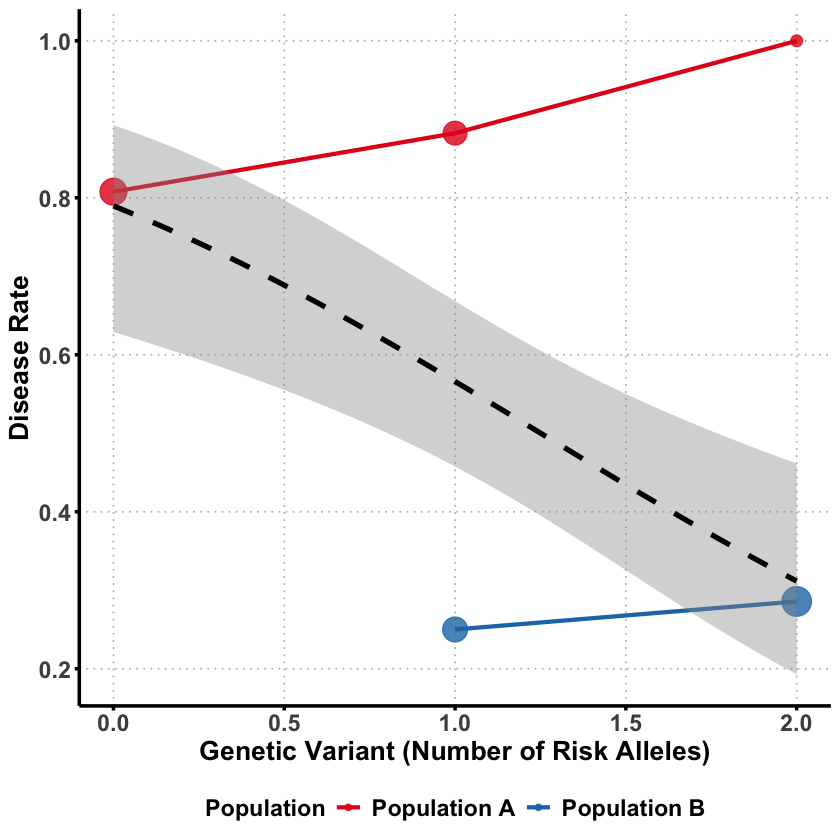
Sometimes a genetic variant can appear protective when analyzed alone, but turn out to be detrimental when we account for other factors. This counterintuitive phenomenon is Simpson’s paradox - where the direction of association completely reverses between marginal and joint analysis.
The key question: How can a genetic variant seem protective overall but actually increase risk when we control for confounding factors?
We’ll simulate a scenario where genetic ancestry acts as a confounder:
rm(list = ls())
set.seed(9) # For reproducibility
N <- 100 # Sample size
# Create a confounding variable (genetic ancestry)
ancestry <- rbinom(N, 1, 0.5) # 0 = Population A, 1 = Population B
Different populations have different allele frequencies and Population B has much higher frequency of the “risk” allele.
# Generate genotype that's correlated with ancestry
# Population B has higher frequency of risk allele
variant1 <- ifelse(ancestry == 0,
rbinom(sum(ancestry == 0), 2, 0.2), # Pop A: low risk allele frequency
rbinom(sum(ancestry == 1), 2, 0.8)) # Pop B: high risk allele frequency
# Check allele frequencies by population
cat("Population A (ancestry=0) mean genotype:", round(mean(variant1[ancestry == 0]), 2), "\n")
cat("Population B (ancestry=1) mean genotype:", round(mean(variant1[ancestry == 1]), 2), "\n")
Population A (ancestry=0) mean genotype: 0.41
Population B (ancestry=1) mean genotype: 1.59
Then we generate the disease status where despite having more risk alleles, Population B has much lower disease rates overall.
# Population B has generally lower disease risk (better healthcare/environment)
# But the variant increases risk within each population
baseline_risk <- ifelse(ancestry == 0, 0.8, 0.1) # Pop A much higher baseline risk
genetic_effect <- 0.1 * variant1 # Variant increases risk in both populations
disease_prob <- baseline_risk + genetic_effect
disease <- rbinom(N, 1, pmin(disease_prob, 1)) # Ensure prob ≤ 1
# Create data frame
data <- data.frame(
disease = disease,
variant1 = variant1,
ancestry = ancestry
)
# Check disease rates by population
cat("Population A disease rate:", round(mean(data$disease[data$ancestry == 0]), 2), "\n")
cat("Population B disease rate:", round(mean(data$disease[data$ancestry == 1]), 2), "\n")
Population A disease rate: 0.81
Population B disease rate: 0.26
First, let’s see what happens when we analyze each population separately:
# Separate data by ancestry
data_ancestry0 <- data[data$ancestry == 0, ] # Population A
data_ancestry1 <- data[data$ancestry == 1, ] # Population B
# Marginal analysis for Population A
marginal_model_A <- glm(disease ~ variant1, data = data_ancestry0, family = binomial)
marginal_OR_A <- exp(coef(marginal_model_A)[2])
# Marginal analysis for Population B
marginal_model_B <- glm(disease ~ variant1, data = data_ancestry1, family = binomial)
marginal_OR_B <- exp(coef(marginal_model_B)[2])
cat("=== WITHIN-POPULATION EFFECTS ===\n")
cat("Population A: OR =", round(marginal_OR_A, 3), "\n")
cat("Population B: OR =", round(marginal_OR_B, 3), "\n")
=== WITHIN-POPULATION EFFECTS ===
Population A: OR = 2.317
Population B: OR = 0.709
Then we analyze the data with two ancestries combined:
# Marginal analysis (ignoring ancestry - combining both populations)
marginal_model <- glm(disease ~ variant1, data = data, family = binomial)
marginal_OR <- exp(coef(marginal_model)[2])
marginal_p <- summary(marginal_model)$coefficients[2, 4]
cat("=== MARGINAL EFFECT (combining populations, ignoring ancestry) ===\n")
cat("OR =", round(marginal_OR, 3), ", p =", round(marginal_p, 4), "\n")
cat("Interpretation:", ifelse(marginal_OR > 1, "Detrimental ", "Protective"), "\n")
=== MARGINAL EFFECT (combining populations, ignoring ancestry) ===
OR = 0.394 , p = 4e-04
Interpretation: Protective
Even though the variant is detrimental in both populations, it appears protective when we ignore ancestry!
Why this happens?
Population B has higher frequency of risk alleles BUT lower baseline disease risk
Population A has lower frequency of risk alleles BUT higher baseline disease risk
Marginally: The variant appears protective because it’s more common in the healthier population
Within each population: The variant actually increases disease risk
Supplementary#
Graphical Summary#
library(ggplot2)
library(dplyr)
# Use the same data from our example
set.seed(42)
N <- 100
ancestry <- rbinom(N, 1, 0.5)
variant1 <- ifelse(ancestry == 0,
rbinom(sum(ancestry == 0), 2, 0.2),
rbinom(sum(ancestry == 1), 2, 0.8))
baseline_risk <- ifelse(ancestry == 0, 0.8, 0.1)
genetic_effect <- 0.1 * variant1
disease_prob <- baseline_risk + genetic_effect
disease <- rbinom(N, 1, pmin(disease_prob, 1))
# Create data frame with proper labels
plot_data <- data.frame(
variant = variant1,
disease = disease,
ancestry = factor(ancestry, levels = c(0, 1), labels = c("Population A", "Population B"))
)
# Calculate summary statistics for each group
summary_stats <- plot_data %>%
group_by(ancestry, variant) %>%
summarise(
disease_rate = mean(disease),
n = n(),
.groups = 'drop'
)
# Create the plot
p <- ggplot(summary_stats, aes(x = variant, y = disease_rate)) +
# Points for each ancestry group
geom_point(aes(color = ancestry, size = n), alpha = 0.8) +
geom_line(aes(color = ancestry, group = ancestry), linewidth = 1.2) +
# Overall trend line (ignoring ancestry)
geom_smooth(data = plot_data, aes(x = variant, y = disease),
method = "glm", method.args = list(family = "binomial"),
se = TRUE, color = "black", linetype = "dashed", linewidth = 1.5) +
scale_color_manual(values = c("Population A" = "#E31A1C", "Population B" = "#1F78B4")) +
scale_size_continuous(range = c(3, 8), guide = "none") +
labs(
x = "Genetic Variant (Number of Risk Alleles)",
y = "Disease Rate",
color = "Population"
) +
theme_minimal() +
theme(
text = element_text(size = 14, face = "bold"),
plot.title = element_blank(),
axis.text = element_text(size = 14, face = "bold"),
axis.title = element_text(size = 16, face = "bold"),
legend.text = element_text(size = 14, face = "bold"),
legend.position = "bottom", # Position legend at the bottom
panel.grid.major = element_line(color = "gray", linetype = "dotted"),
panel.grid.minor = element_blank(),
axis.line = element_line(linewidth = 1),
axis.ticks = element_line(size = 1),
# Add transparent backgrounds
panel.background = element_rect(fill = "transparent", color = NA),
plot.background = element_rect(fill = "transparent", color = NA)
)
# Display the plot
print(p)
# Save with transparent background
ggsave("./cartoons/marginal_joint_effects.png", plot = p,
width = 8, height = 8,
bg = "transparent",
dpi = 300)