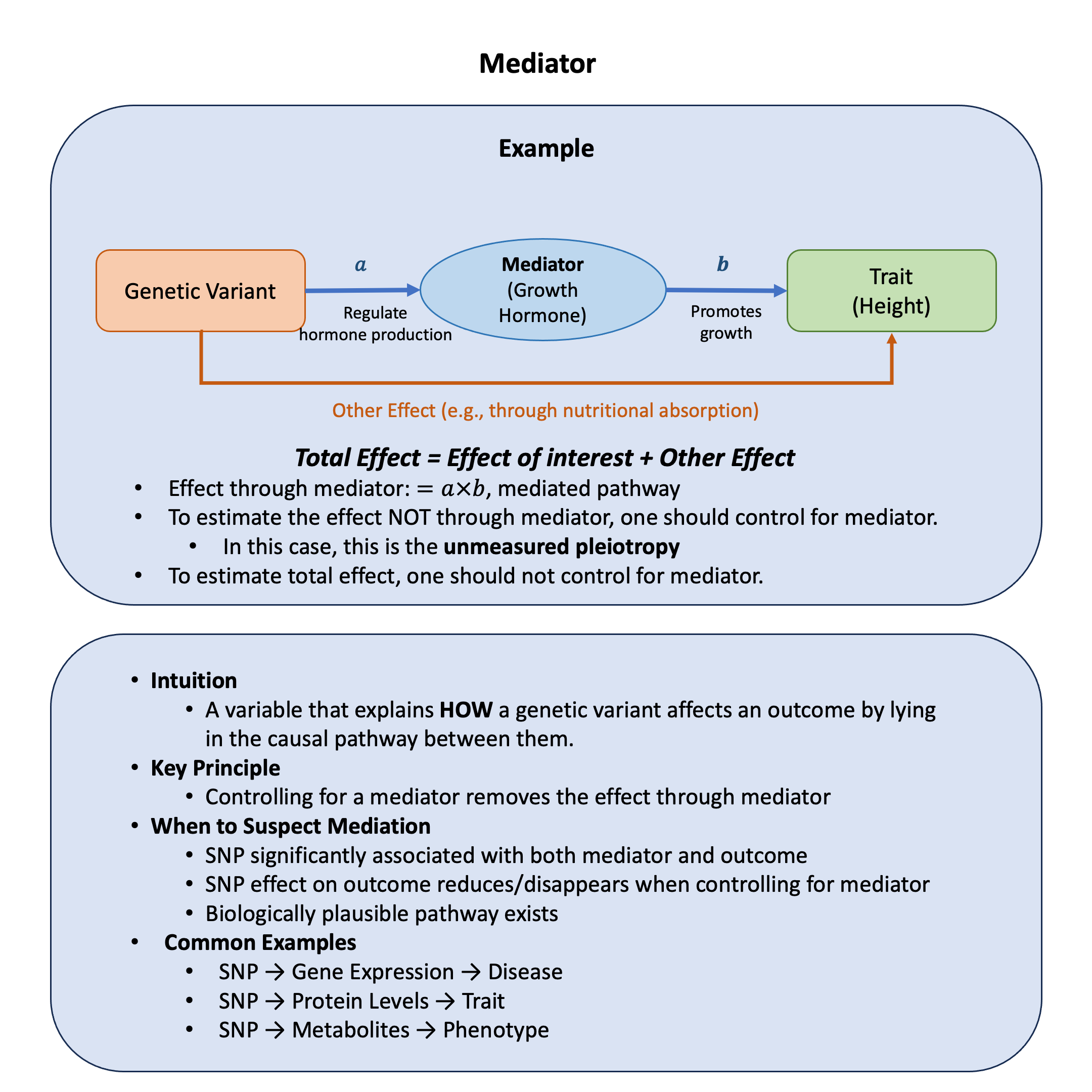
Mediator#
A mediator is a variable that sits in the causal pathway between an exposure and an outcome, explaining the mechanism through which the exposure exerts its effect on the outcome.
Graphical Summary#
Key Formula#
The key formula for the concept of a mediator is represented in a causal diagram as:
Where:
\(X\) is the independent variable (e.g., genetic variant)
\(W\) is the mediator variable
\(Y\) is the dependent variable (e.g., trait)
The arrows (\(\rightarrow\)) indicate the direction of causal influence
This diagram illustrates that a mediator (\(W\)) lies in the causal pathway between the independent variable (\(X\)) and the dependent variable (\(Y\)). The mediator transmits the effect of \(X\) on \(Y\), creating a causal pathway through which \(X\) affects \(Y\).
Technical Details#
What is Mediation?#
A mediator is a variable that explains the mechanism by which a genetic variant affects an outcome. Unlike confounders (which create bias) or colliders (which induce bias when controlled), mediators represent the actual biological pathway.
Mediation vs Other Variable Types#
When deciding whether to control for a variable, ask:
Mediator: “Does this variable explain HOW the SNP affects the outcome?”
Action: Can control to isolate effects not through the mediator
Structure: SNP -> Mediator -> Outcome
Confounder: “Does this variable affect both SNP and outcome?”
Action: Must control to remove bias
Structure: SNP <- Confounder -> Outcome
Collider: “Is this variable caused by both SNP and outcome?”
Action: Never control - creates bias
Structure: SNP -> Collider <- Outcome
The Mediation Framework#
Where:
Total Effect: SNP -> Outcome (\(\beta\) without controlling for mediator)
Effect through Mediator: SNP -> Mediator -> Outcome (the mediated pathway = \(a \times b\))
Other Effects: Effect NOT through the mediator (\(\beta\) when controlling for mediator - includes unmeasured pleiotropy)
Evidence for Mediation#
Strong evidence when controlling for the mediator:
Reduces effect size: Total effect > Effect after controlling for mediator
Eliminates significance: p-value increases substantially
Biological plausibility: Mediator is in known pathway
Analysis Steps#
Estimate total effect:
lm(Outcome ~ SNP)(should be significant)Test for mediation:
lm(Outcome ~ SNP + Mediator)(SNP effect should reduce/disappear)
Interpretation:
If SNP effect disappears -> Complete mediation
If SNP effect reduces -> Partial mediation
If SNP effect unchanged -> No mediation
Examples of Mediation:
SNP -> Gene Expression -> Disease
SNP -> Protein Levels -> Metabolic Trait
SNP -> Hormone Levels -> Growth/Development
SNP -> Enzyme Activity -> Drug Response
Example#
Imagine you discover that a genetic variant is associated with height. The natural follow-up question is: How does this variant actually influence height? Does it work through growth hormones, bone development, or some other biological pathway?
Understanding the mechanism matters because it tells us where and how we might intervene. If a variant affects height through growth hormone levels, then measuring growth hormone could help us understand individual differences in height and potentially guide treatment decisions.
Here’s our classic scenario: We have 5 individuals with different genetic variants, and we want to understand how one particular variant influences height. We suspect that growth hormone might be the key mediator - the biological “middleman” that explains how genes influence height.
The crucial question is: What happens to the genetic association with height when we account for growth hormone levels? Does the genetic effect disappear (suggesting complete mediation) or remain (suggesting other pathways)?
The crucial question is: If growth hormone is truly the only mechanism by which this variant affects height, then controlling for growth hormone levels should remove the genetic association with height. Why? Because if this is the only pathway, then once we account for the mediator, there should be no remaining pathway for the variant to influence height.
# Clear the environment
rm(list = ls())
set.seed(16)
# Define genotypes for 5 individuals at 3 variants
# These represent actual alleles at each position
# For example, Individual 1 has genotypes: CC, CT, AT
genotypes <- c(
"CC", "CT", "AT", # Individual 1
"TT", "TT", "AA", # Individual 2
"CT", "CT", "AA", # Individual 3
"CC", "TT", "AA", # Individual 4
"CC", "CC", "TT" # Individual 5
)
# Reshape into a matrix
N = 5
M = 3
geno_matrix <- matrix(genotypes, nrow = N, ncol = M, byrow = TRUE)
rownames(geno_matrix) <- paste("Individual", 1:N)
colnames(geno_matrix) <- paste("Variant", 1:M)
alt_alleles <- c("T", "C", "T")
# Convert to raw genotype matrix using the additive / dominant / recessive model
Xraw_additive <- matrix(0, nrow = N, ncol = M) # dount number of non-reference alleles
rownames(Xraw_additive) <- rownames(geno_matrix)
colnames(Xraw_additive) <- colnames(geno_matrix)
for (i in 1:N) {
for (j in 1:M) {
alleles <- strsplit(geno_matrix[i,j], "")[[1]]
Xraw_additive[i,j] <- sum(alleles == alt_alleles[j])
}
}
X <- scale(Xraw_additive, center = TRUE, scale = TRUE)
We assign the growth hormones levels for each individual from variant 3:
# Generate growth hormone levels FROM variant 3 (mediator pathway)
GH_raw <- 6 + 2 * Xraw_additive[, 3] + rnorm(N, 0, 0.1) # Variant 3 affects GH
GH <- scale(GH_raw)
Then we assign the height for the individuals (mediated by hormones):
# Create mediator structure: Variant 3 -> Growth Hormone -> Height
# Height is caused by:
# 1. Direct effect from growth hormone (the mediator)
# 2. Small effects from variants 1 and 2 (not mediated)
# 3. NO direct effect from variant 3 (fully mediated through GH)
height_raw <- 160 + # Base height
3 * GH + # Growth hormone effect (mediator pathway)
1 * X[, 1] + # Small direct effect from variant 1
0.5 * X[, 2] + # Small direct effect from variant 2
0 * X[, 3] + # NO direct effect from variant 3 (fully mediated)
rnorm(N, 0, 0.5) # Small noise
Y <- scale(height_raw)
Then we perform OLS regression for each SNP:
p_values <- numeric(M) # Store p-values
betas <- numeric(M) # Store estimated effect sizes
p_values_adjusted <- numeric(M) # Store p-values adjusted for GH
betas_adjusted <- numeric(M) # Store estimated effect sizes adjusted for GH
# Perform OLS regression for each SNP
for (j in 1:M) {
SNP <- X[, j] # Extract genotype for SNP j
model <- lm(Y ~ SNP) # OLS regression: Trait ~ SNP
adjusted_model <- lm(Y ~ SNP + GH) # Adjust for GH
summary_model <- summary(model)
summary_adjusted_model <- summary(adjusted_model)
# Store p-value and effect size (coefficient)
p_values[j] <- summary_model$coefficients[2, 4] # p-value for SNP effect
betas[j] <- summary_model$coefficients[2, 1] # Estimated beta coefficient
p_values_adjusted[j] <- summary_adjusted_model$coefficients[2, 4] # p-value for SNP effect adjusted for growth hormone
betas_adjusted[j] <- summary_adjusted_model$coefficients[2, 1] # Estimated beta coefficient adjusted for growth hormone
}
# Create results table
results <- data.frame(Variant = colnames(X), Beta = betas, P_Value = p_values,
Beta_Adjusted = betas_adjusted, P_Value_Adjusted = p_values_adjusted)
results
| Variant | Beta | P_Value | Beta_Adjusted | P_Value_Adjusted |
|---|---|---|---|---|
| <chr> | <dbl> | <dbl> | <dbl> | <dbl> |
| Variant 1 | -0.3763359 | 0.532398192 | 0.2151202 | 0.1845598 |
| Variant 2 | 0.9133526 | 0.030216252 | 0.2247750 | 0.5336030 |
| Variant 3 | 0.9668181 | 0.007219589 | -3.0237908 | 0.3374487 |
The results perfectly demonstrate the concept of mediation in genetic association studies. In the total effect analysis (without controlling for growth hormone), Variant 3 shows significant associations with height (p = 0.007). However, when we control for growth hormone as a mediator, the pattern dramatically changes: Variant 3’s association completely disappears (p-value increases from 0.007 to 0.337), indicating that its effect on height is entirely through the growth hormone pathway rather than through other mechanisms.
This illustrates the key principle that controlling for a mediator reveals the effect NOT through that mediator. For Variant 3, the loss of significance after controlling for growth hormone confirms that this variant affects height specifically through its influence on growth hormone levels, with no additional pathways contributing to the association. This represents a classic complete mediation scenario in genetic studies.